Installation of pyvo
In order to interact with the TAP interface of gaia.aip.de you only require
python 3+ and pyvo 1+.
pip install pyvo>=1.0
Importing PyVo and checking the version
It is useful to always print the version of pyvo you are using. Most of non-working scripts fail because of an old version of pyvo.
from pkg_resources import parse_version
import pyvo
#
# Verify the version of pyvo
#
if parse_version(pyvo.__version__) < parse_version('1.0'):
raise ImportError('pyvo version must be at least than 1.0')
print('\npyvo version %s \n' % (pyvo.__version__,))
Authentication
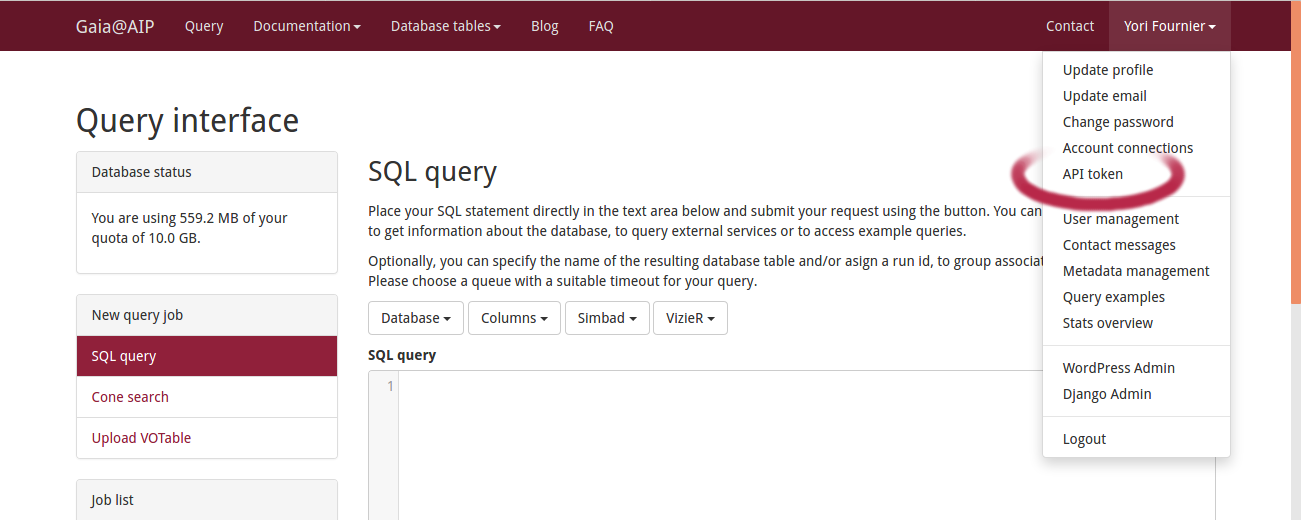
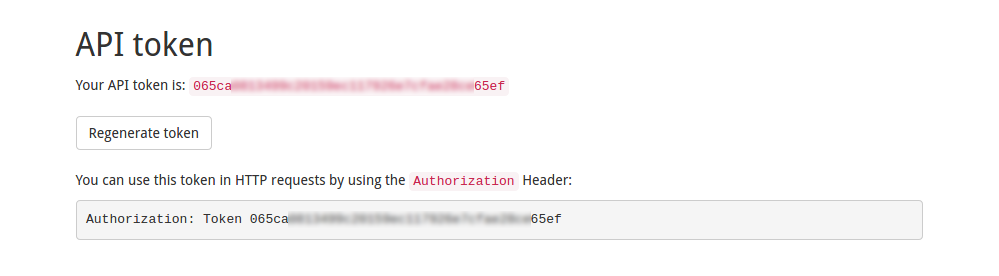
After registration you can access your API Token by clicking on your user name in the right side of the menu bar. Then select API Token.
You will see a long alphanumerical word. Just copy it where ever you see <your-token> ; in the following examples.
The
API Tokenidentifies you and provides access to the results tables of your queries.
The connection to the TAP service can be done that way:
import requests
import pyvo
#
# Setup tap_service connection
#
service_name = "Gaia@AIP"
url = "https://gaia.aip.de/tap"
token = 'Token <your-token>'
print('TAP service %s \n' % (service_name,))
# Setup authorization
tap_session = requests.Session()
tap_session.headers['Authorization'] = token
tap_service = pyvo.dal.TAPService(url, session=tap_session)
Short queries
Many queries last less than a few seconds, we call them short queries. The latter can be executed with synchronized jobs. You will retrieve the results interactively.
lang = "PostgreSQL"
query = '''
-- Orbital elements of Solar System Objects
SELECT osc_epoch, orb_m, omega, node_omega, inclination, eccentricity, a
FROM gaiadr2.aux_sso_orbits;
'''
tap_result = tap_service.run_sync(query, language=lang)
Remark: the
langparameter can take two values eitherPostgreSQLorADQLthis allows to access some featured present in the one or the other language for more details about the difference between both please refer : ADQL or to IVOA docs
The result tap_result is a so called TAPResults that is essentially a wrapper around an Astropy votable.Table.
Asynchronous jobs
For slightly longer queries, typically counting or larger selections (>10000 objects), a synchronized job will fail because of timeouts (from http protocol or server settings). This is why we provide the possibility to submit asynchronous jobs. These type of jobs will run on the server side, store their results so that you can retrieve them at a later time. Choose one of the 3 queues:
- 30 second queue
- 5 min queue
- 2 hours queue
The 30 seconds queue
Most of the asynchronous queries will require less than 30 seconds, basically all queries without JOIN, or CONE SEARCH. Therefore this queue is the default and should be preferred.
#
# Submit the query as an async job
#
query_name = "tgas_stars"
lang = 'PostgreSQL' # ADQL or PostgreSQL
query = '''
-- Number of TGAS stars with parallax / parallax_error > 10
SELECT COUNT(*)
FROM gaiadr1.tgas_source
WHERE parallax / parallax_error > 10;
'''
job = tap_service.submit_job(query, language=lang, runid=query_name, queue="30s")
job.run()
#
# Wait to be completed (or an error occurs)
#
job.wait(phases=["COMPLETED", "ERROR", "ABORTED"], timeout=30.0)
print('JOB %s: %s' % (job.job.runid, job.phase))
#
# Fetch the results
#
job.raise_if_error()
print('\nfetching the results...')
tap_results = job.fetch_result()
print('...DONE\n')
As for sync jobs, the result is a TAPResults object.
The 5 minutes queue
If you want to extract information on specific stars from various tables you have to JOIN tables. Your query may need more than a few seconds. For that, the 5 minutes queue provides a good balance. It should be noticed that for such a queue the wait method should not be used to prevent an overload of the server at peak usage. Therefore using the script with the sleep() method is recommended.
import time
#
# Submit the query as an async job
#
lang = 'ADQL'
query_name = "glob_cluster"
query = '''
-- M4 globular cluster with geometric distances using ADQL
SELECT gaia.source_id, gaia.ra, gaia.dec, gd.r_est
FROM gaiadr2.gaia_source gaia, gaiadr2_contrib.geometric_distance gd
WHERE 1 = CONTAINS(POINT('ICRS', gaia.ra, gaia.dec),
CIRCLE('ICRS',245.8962, -26.5222, 0.5))
AND gaia.phot_g_mean_mag < 15
AND gd.r_est BETWEEN 1500 AND 2300
AND gaia.source_id = gd.source_id
'''
job = tap_service.submit_job(query, language=lang, runid=query_name, queue="5m")
job.run()
print('JOB %s: SUBMITTED' % (job.job.runid,))
#
# Wait for the query to finish
#
while job.phase not in ("COMPLETED", "ERROR", "ABORTED"):
print('WAITING...')
time.sleep(120.0) # do nothing for some time
print('JOB ' + (job.phase))
#
# Fetch the results
#
job.raise_if_error()
print('\nfetching the results...')
results = job.fetch_result()
print('...DONE\n')
The 2 hours queue
Some complex queries like Cross-Matching or geometric search may take more than the short queues allow. For this purpose we provide the 2 hours queue and the 24 hours queue (only on request). If you need longer queue times please contact us.
When running a long query, you surely don't want to block CPU ressources for a python process that just waits for 2 hours, for the queue to finish. Therefore, long queries are typically done in two parts (= two scripts), one that submits the request, another one that retrieves the results.
Submitting a job and store job_urls for later retrieval
We first submit the query as an async job to the 2h queue, and store the job (the url) of the newly created job into a file job_url.txt. With this url we are able to retrieve the results (when it has finished) at any later time.
#
# Submit the query as an async job
#
query_name = "glob_cluster"
lang = 'PostgreSQL'
query = '''
-- M4 globular cluster using PostgreSQL
SELECT ra, dec, phot_g_mean_mag AS gmag
FROM gaiadr1.gaia_source
WHERE pos @ scircle(spoint(RADIANS(245.8962), RADIANS(-26.5222)), RADIANS(0.5))
AND phot_g_mean_mag < 15
'''
job = tap_service.submit_job(query, language=lang, runid=query_name, queue="2h")
job.run()
print('JOB %s: SUBMITTED' % (job.job.runid,))
print('JOB %s: %s' % (job.job.runid, job.phase))
#
# Save the job's url in a file to later retrieve results.
#
print('URL: %s' % (job.url,))
with open('job_url.txt', 'w') as fd:
fd.write(job.url)
Retrieve the results at a later time
In order to retrieve the results, we will first recreate the job from the job_url stored in the job_url.txt file and verify that the job is finished, by asking for its current phase. In case the job is finished we will retrieve the results as usual.
#
# Recreate the job from url and session (token)
#
# read the url
with open('job_url.txt', 'r') as fd:
job_url = fd.readline()
# recreate the job
job = pyvo.dal.AsyncTAPJob(job_url, session=tap_session)
#
# Check the job status
#
print('JOB %s: %s' % (job.job.runid, job.phase))
# if still running --> exit
if job.phase not in ("COMPLETED", "ERROR", "ABORTED"):
exit(0)
#
# Fetch the results
#
job.raise_if_error()
print('\nfetching the results...')
tap_results = job.fetch_result()
print('\n...DONE\n')
Thanks to this method you can submit a job, go for a coffee, write a paper and retrieve the results when it suits you. The job and its results are stored on the server side under your user account.
Submitting multiple queries
Some time it is needed to submit several queries at one time. Either because the entire query may last longer than 2 hours and you need to cut it in smaller parts, or because you need non JOIN-able information from various tables.
Your query is too long? Chunk it!
Before contacting us and ask for longer queue time: You may try to cut the long query into chunks, and execute your long query as a list of shorter queries.
There are three typical ways to do it:
- chunk via random index
- chunk via sky position: (ra, dec)
- chunk via healpix
List of file queries
Sometimes it is useful to just send all .sql queries present in a directory. For such purpose you can use comments to provide the proper parameters.
Let us consider the file glob_cluster.sql
-- M4 globular cluster with geometric distances using ADQL
-- LANGUAGE = ADQL
-- QUEUE = 5m
SELECT gaia.source_id, gaia.ra, gaia.dec, gd.r_est
FROM gaiadr2.gaia_source gaia, gaiadr2_contrib.geometric_distance gd
WHERE 1 = CONTAINS(POINT('ICRS', gaia.ra, gaia.dec),
CIRCLE('ICRS',245.8962, -26.5222, 0.5))
AND gaia.phot_g_mean_mag < 15
AND gd.r_est BETWEEN 1500 AND 2300
AND gaia.source_id = gd.source_id
The language and queue are prescibed as comments. The query can then be submitted in a script like the following:
import glob
#
# Submit the query as an Asynchrone job
#
# find all .sql files in current directory
queries_filename = sorted(glob.glob('./*.sql'))
print('Sending %d examples' % (len(queries_filename),))
# initialize test results
jobs = []
failed = []
# send all queries
for query_filename in queries_filename:
# read the .SQL file
with open(query_filename, 'r') as fd:
query = ' '.join(fd.readlines())
# Set language from comments (default: PostgreSQL)
if 'LANGUAGE = ADQL' in query:
lang = 'ADQL'
else:
lang = 'PostgreSQL'
# Set queue from comments (default: 30s)
if 'QUEUE = 5m' in query:
queue = "5m"
elif 'QUEUE = 2h' in query:
queue = "2h"
else:
queue = "30s"
# Set the runid from sql filename
base = os.path.basename(query_filename)
runid = os.path.splitext(base)[0]
print('\n> Query : %s\n%s\n' % (runid, query))
Cone search examples
One of the great feature of the ADQL language is to provide spatial and geometrical access.
This allows Gaia@AIP to provide a Cone Search feature. The latter is accessible via the web interface in the Cone Search section of the query menu. However it is also possible to request Cone Search from the TAP interface using pyvo.
Let us consider the M4 globular cluster: its coordinates are (245.8962, -26.5222) in degrees with a search radius of 0.5 arcsec.
Requesting all sources found in the globular cluster could look like:
#
# Setting the cone search parameters
#
# RA (in degrees)
ra = 245.8962
# DEC (in degreees)
dec = -26.5222
# Radius (in degrees)
sr = 0.5
#
# Submit the query as an async job
#
query_name = "simple_cs_adql"
lang = 'ADQL'
query = '''
-- Simple cone search with ADQL
SELECT ra, dec, DISTANCE( POINT('ICRS', ra, dec), POINT('ICRS', %(ra)f, %(dec)f) ) AS dist
FROM gaiadr3.gaia_source
WHERE 1 = CONTAINS( POINT('ICRS', ra, dec), CIRCLE('ICRS', %(ra)f, %(dec)f, %(radius)f) );
''' % {"ra": ra, "dec": dec, "radius": sr}
job = tap_service.submit_job(query, language=lang, runid=query_name, queue="2h")
job.run()
The pgSphere extension of PostgreSQL provides a similar interface to spatial and geometrical searches allowing for Cone Search.
At Gaia@AIP we provide such an interface and accessing Cone Search with PostgreSQL can be done as:
#
# Setting the cone search parameters
#
# RA (in degrees)
ra = 245.8962
# DEC (in degrees)
dec = -26.5222
# Radius (in degrees)
sr = 0.5
#
# Submit the query as an async job
#
query_name = "simple_cs_pg"
lang = 'PostgreSQL'
query = '''
-- Simple cone search with PostgreSQL
-- WARNING: dist is in radians
SELECT ra, dec, spoint(RADIANS(ra), RADIANS(dec)) <-> spoint(RADIANS(%(ra)f), RADIANS(%(dec)f)) AS dist
FROM gaiadr3.gaia_source
WHERE pos @ scircle(spoint(RADIANS(%(ra)f), RADIANS(%(dec)f)), RADIANS(%(radius)f));
''' % {"ra": ra, "dec": dec, "radius": sr}
job = tap_service.submit_job(query, language=lang, runid=query_name, queue="2h")
job.run()
Convert result to various python types
The results obtained via the fetch_results() method returns a so called TAPResults object. The latter is essencially a votable. In case you are not familiar with votables here is a few tricks to get back to some more general pythonic types.
-
Print the data:
python tap_results.to_table().pprint(max_lines=10)It is important to notice themax_lineskeyword, printing too many lines may crash a low-memory machine. -
Show as html (in a browser):
python tap_results.to_table().show_in_browser(max_lines=10)It is important to notice themax_lineskeyword, printing too many lines may crash a low-memory machine. -
Show in a notebook (ipython, jupyter or jupyterlab):
python tap_results.to_table().show_in_notebook(display_length=10)It is important to notice thedisplay_lengthkeyword, printing too many lines may crash a low-memory machine. -
Get a numpy array:
python np_array = tap_results.to_table().as_array() -
Get a Panda's DataFrame
python df = tap_results.to_table().to_pandas()- Get the header of DataFrame:
python df.head()
- Get the header of DataFrame:
Archiving your jobs
If you submit several large queries you may go over quota: set to 10 GB. In order to avoid to get over quota you may consider archiving your jobs. Archiving removes the data from the server side but keeps the SQL query. This allows to resubmit a query at a later time.
Deleting (Archiving) a job with pyvo can be simply done that way:
job.delete()
Archiving all COMPLETED jobs
A nice feature of the TAP service is to retrieve all jobs that are marked as COMPLETED and archive them at ones. This can be done as follows:
#
# Archiving all COMPLETED jobs
#
# obtain the list of completed job_descriptions
completed_job_descriptions = tap_service.get_job_list(phases='COMPLETED')
# Archiving each of them
for job_description in completed_job_descriptions:
# get the jobid
jobid = job_description.jobid
# recreate the url by hand
job_url = tap_service.baseurl + '/async/' + jobid
# recreate the job
job = pyvo.dal.AsyncTAPJob(job_url, session=tap_session)
print('Archiving: {url}'.format(url=job_url))
job.delete() # archive job
Rerunning ARCHIVED jobs
Rerunning and retrieving results from a job that have been archived previously, can be achieved that way:
#
# Re-running Archived jobs
#
# obtain the list of the two last ARCHIVED job_descriptions
archived_job_descriptions = tap_service.get_job_list(phases='ARCHIVED', last=2)
# re-running the two last Archived jobs
for job_description in archived_job_descriptions:
# get jobid
jobid = job_description.jobid
# recreate the url by hand
job_url = tap_service.baseurl + '/async/' + jobid
# recreate the archived job
archived_job = pyvo.dal.AsyncTAPJob(job_url, session=tap_session)
# get the language (with a bit of magic)
lang = [parameter._content for parameter in archived_job._job.parameters if parameter._id == 'query_language'][0]
# extract the query
query = archived_job.query
# resubmit the query with corresponding parameters
job = tap_service.submit_job(query, language=lang, runid='rerun', queue='30s')
print('%(url)s :\n%(query)s\n' % {"url": job_url, "query": query})
# start the job
try:
job.run()
except pyvo.dal.DALServiceError:
raise ValueError("Please check that the SQL query is valid, and that the SQL language is correct.")
Retrieving the results is done as explained above.
If you prefer you can also filter for a given runid.
#
# Filtering by runid
#
target_runid = 'glob_cluster'
# obtain the list of completed job_descriptions
archived_job_descriptions = tap_service.get_job_list(phases='ARCHIVED')
for job_description in archived_job_descriptions:
# select the job with runid fitting target_runid
if job_description.runid == target_runid:
# get jobid
jobid = job_description.jobid
# recreate the url by hand
job_url = tap_service.baseurl + '/async/' + jobid
# recreate the archived job
archived_job = pyvo.dal.AsyncTAPJob(job_url, session=tap_session)
# get the language (with a bit of magic)
lang = [parameter._content for parameter in archived_job._job.parameters if parameter._id == 'query_language'][0]
# extract the query
query = archived_job.query
# resubmit the query with corresponding parameters
job = tap_service.submit_job(query, language=lang, runid='rerun', queue='30s')
print('%(url)s :\n%(query)s\n' % {"url": job_url, "query": query})
# start the job
try:
job.run()
except pyvo.dal.DALServiceError:
raise ValueError("Please check that the SQL query is valid, and that the SQL language is correct.")